Matrix-assisted laser desorption/ionization time-of-flight mass spectrometry for the identification of beta-hemolytic streptococci
The application of matrix-assisted laser desorption/ionization time-of-flight mass spectrometry (MALDI-TOF MS) has greatly facilitated the microbial isolate identification in clinical microbiology laboratories, most notably by drastically decreasing the turn-around-time in comparison to the conventional methods. Other key advantages offered by MALDI-TOF MS include improved sensitivity, accuracy and resolution power in identifying microbial isolates (1). The technique has been widely used for the identification of bacteria, mycobacteria and fungi, and the accuracy varied by species (1-5). Early studies evaluated the possibility of intact cell identification by MALDI-TOF MS, suggesting the capability to eliminate protein extraction prior to strain analysis, which has become a standard practice for clinical bacteria identification (6,7).
Beta-hemolytic streptococci (BHS) are a family of streptococci species composed of different serogroups, including Lancefield antigenic group A (S. pyogens), antigenic group B (S. agalactiae), antigenic group C (S. dysgalactiae among others), and Group G (S. canis among others). The group of BHS causes a wide variety of diseases, ranging from mild pharygitis to life threatening sepsis. Therefore, timely identification of these bacteria, especially for more critical important pathogens such as S. pyogens and S. agalactiae, would greatly enhance patient care and decrease disease burden. Early studies using MALDI-TOF MS to characterize the intact-cell spectra generated from ionization of intact cells of three major groups of BHS before automated databases were available, and they were already able to detect differences in the spectra of group A, C and D streptococci with excellent reproducibility (8). In our study, we evaluate the performance of two commercial available MALDI-TOF MS systems: VITEK MS system (bioMerieux, Marcy l’Etoile, France) and the MALDI BioTyper system (Bruker Daltonics, Bremen, Germany), in identifying clinical isolates of BHS, and compared to that of the traditional BD Phoenix automated system.
Materials and methods
Bacterial isolates
All strains used in this study were isolated from clinical sources including blood, urine, pus, sterile body fluid and respiratory specimens in Zhongshan Hospital of Fudan University in Shanghai, China. Clinical samples were inoculated on 5% sheep blood Columbia agar (bioMerieux, Marcy l’Etoile) for 18 to 24 h at 35 °C with 5% CO2. According to characteristic colony appearance, biochemical results, and agglutination reaction, BHS were isolated and identified using BD Phoenix SMIC/ID streptococcal panels (Becton Dickinson, city, United States), VITEK MS bioMerieux system and BioTyper MS system respectively.
BD phoenix identification
Isolates were inoculated within BD Phoenix SMIC/ID streptococcal panels (BD PhoenixSID, panel448505) with a density of 0.5 McF. Once inoculated, panels were placed into the BD Phoenix Automated Microbiology System (Becton Dickinson City, United States) and incubated at 35 °C. According to the manufacturer’s recommendations, identification with a score <90% in the BD Phoenix system was thresholded as “no result”. In cases of “no result”, the identifications were repeated using colonies inoculated for up to 48 h.
VITEK MS identification
Target slide sample preparation, mass spectra acquisition, and mass spectra profile processing were performed with the VITEK MS bioMerieux system using an Axima Assurance mass spectrometer with the acquisition software Version 1.1 and the IVD database Version 2.0. According to the manufacturer’s recommendations, a confidence value between 60-99.9% with a single species proposed was considered to be accurate. If a unique ID pattern was not recognized comparison to the database, a list of possible organisms was provided. The result was classified as a low-discrimination (LD) when a confidence value was >60% for each proposed species. In cases with confidence values <60%, the strain was determined to be outside of the scope of the database and the results were classified as no identification (no ID). In cases of poor-quality spectra LD results, and no-ID results, a new acquisition run was performed. When required, the isolates were re-tested using a new single deposit.
BioTyper MS identification
The MicroFlex MALDI-TOF mass spectrometer with MALDI Biotyper software 2.0 (Bruker Daltonics, Bremen, Germany) was used for strain identification. Sample extractions and strain identifications were performed following the manufacturer’s instruction. A score of >2 indicated secure genus and probable species identification. For isolates with a score <2, the test was repeated. If the repeat testing still resulted in a score <2 but ≥1.7, the score was recorded with the result.
In circumstances of discordant identification results between VITEK MS, BioTyper MS and BD Phoenix, 16sRNA sequencing of the strains was performed to obtain the reference ID.
16S rRNA identification
Bacterial DNA was extracted from inoculated colonies after heating at 95 °C for 10 min. DNA extracts and controls were amplified using DreamTaq Green PCR Master Mix (Thermo Scientific, United States). Amplification conditions were as follow: 5 min at 95 °C to denature the DNA, followed by 30 cycles of denaturation at 94 °C for 30 secs, primer annealing at 55 °C for 30 secs and extension at 72 °C for 30 secs on a Veriti™ 96 thermal cycler (ABI, Vernon, USA). Primers used for 16sRNA amplification were as follow: forward, 5'- TGGAGAGTTTGATCCTGGCTCAG - 3'; reverse, 5'- TACCGCGGCTGCTGGCAC - 3'. Purified PCR products were sequenced in the forward and reverse direction in separate reactions and in duplicates using services provided by BGI Tech Solutions (BGI-Tech, China). Strains were identified by comparing consensus sequences in the GenBank database (http://www.ncbinlm.nih.gov/blast/Blast.cgi) through multiple sequence alignment.
Results
A total of 96 isolates of BHS were analyzed. Of these 96 isolates, 43 isolates of S. dysgalactiae, 21 isolates of S. agalactiae, 18 isolates of S. pyogenes, 9 isolates of S. constellatus, 3 isolates of S. acidmininus, 1 isolate of S. procinus, and 1 isolate of S. anginosus was identified by the BD Phoenix system. Thirty-six isolates, including 14 S. dysgalactiae strains, 3 S. agalactiae, 12 S. pyogenes, 4 S. constellatus, 1 S. acidomininus, 1 S. procinus, and 1 S. anginosus strain yielded “no result” in the first run using colonies inoculated for 24 h. However, when used colonies inoculating for 48 h in the second run, all 36 strains yielded acceptable identification by BD Phoenix. In comparison, the VITEK MS system provided high-quality identifications for all of the strains tested, while the Bruker BioTyper system generated probable species identification for 92 out of the total 96 strains. The four strains with identification score of <2 in the first run were subjected to additional testing, with 3 of them achieving probable species identification, and one S. anginosus retrieving a sub-optimal score of 1.973.
The concordant results of Phoenix, Vitek MS and Biotyper MS were considered as accurate identification in our study, and in case of discordant, 16S rRNA sequence was carried out as the reference identification. In total, when comparing to the 16S rRNA sequencing method, 76 (79.2%), 91 (94.7%), and 92 (95.8%) out of 96 strains were accurately identified at the species level by the Phoenix system, BioTyper MS system, and VITEK MS system, respectively (Table 1).
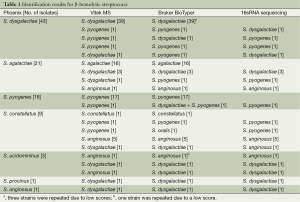
Full table
Discussion
The rapid and accurate identification of pathogens is crucial for clinical practice. However, conventional manual and automated methods are time-consuming and often require complex procedures and large amounts of biological materials. Moreover, conventional phenotypic methods are unable to provide conclusive results in cases of conflicting results between multiple tests (9,10). MALDI-TOF MS has become the new paradigm in providing rapid and accurate bacterial identification results in clinical microbiology laboratories. The efficacy of MALDI-TOF MS to identify bacterial strains at the species level ranges from 100% for a variety of bacteria to about 75% for anaerobic bacteria (1,11,12). Absence of identification or misidentification of the bacteria by MALDI-TOF MS was largely due to the absence of spectra from the databases, or the presence of highly similar spectra between closely related species.
Earlier studies have revealed the capability of MALDI-TOF MS for the accurate identification of streptococcus species. Van Veen et al. accurately identified 85.2% of streptococcal isolates using MALDI-TOF MS at the species level. Misidentifications were associated with viridans group streptococci, in which 12 of 21 (57.1%) viridans group streptococci isolates were falsely identified as S. pneumonia (13). Cherkaoui et al. compared the identification of 386 BHS isolates using MALDI-TOF MS with traditional phenotypic analysis by Vitek-2 system in combination with the latex agglutination test. Using MALDI-TOF MS, all of the isolates were properly identified at the species level with high confidence scores, while only 85% of them were identified with high confidence using phenotypic methods (14). In our study, 96 clinically isolated BHS strains were tested, where 36 isolates needed to be repeated for identification due to unacceptable scores using the BD Phoenix system. In comparison, MALDI-TOF MS provided significantly improved performance, with the VITEK MS system provided complete, accurate identification result with a single run, and the Bruker BioTyper system only needed to repeat the identification for four isolates. Interestingly, using colonies inoculated for 48 h when subjected to re-test by the Phoenix system, all of the 36 strains gained acceptable identification score, highlighting the importance of inoculation duration in traditional phenotypic identification.
Eight of nine isolates misidentified as S. constellatus by the Phoenix system were confirmed as S. pyogenes and S. anginosus using 16S rRNA sequencing. Five of 96 isolates were misidentified using the BioTyper system, four of which were due to trouble distinguishing between S. pyogenes and S. dysgalatiae. Similar results were observed using the VITEK MS system, where 2 of the 3 misidentifications due to difficulty distinguishing S. pyogenes and S. dysgalatiae. One of the strains was identified as mixed species by BioTyper system, one of them was consisted with the 16sRNA sequencing result. It was possibly due to the colony contamination, but also may be caused by distinguishing MS spectrum of S. pyogenes and S. dysgalatiae.
Similar to other reports, our study is not without limits. First, not all isolates were subjected to 16S rRNA sequencing for reference identification. There was a chance of misidentification by all three methods, although the likelihood is quite low. Second, the total number of isolates assayed in this study was relatively small. Further, not all species of BHS were included in this study. However, the isolates analyzed in our study do represent the clinical epidemiology of patients in our health center.
Overall, we conclude that both the BioTyper MS and the VITEK MS were superior methods compared to the conventional phenotypic methods for the identification of BHS. The utility of MALDI-TOF MS in the clinical microbiology laboratory will surely have a positive impact on clinical treatments owing to its rapid and accurate performances of bacterial identification.
Acknowledgements
Authors’ contributions: Chunmei Zhou and Lili Tao contributed equally to study design, data collection, data analysis, and drafting the manuscript. Dr. Bijie Hu contributed to study design, data analysis, critical review, and revision of the manuscript. Jian Ma and Xiangru Ye contributed to collect and analyze data with Chunmei Zhou, Lili Tao and Bijie Hu. Shenglei Huang, Yan Ma and Yuzhang Shan performed these experiments with Chunmei Zhou and Lili Tao, also contributed to data analysis.
Funding: The study was supported by the grant of National Key Clinical Labratory Development Project.
Disclosure: The authors declare no conflict of interest.
References
- Clark AE, Kaleta EJ, Arora A, et al. Matrix-assisted laser desorption ionization-time of flight mass spectrometry: a fundamental shift in the routine practice of clinical microbiology. Clin Microbiol Rev 2013;26:547-603. [PubMed]
- Ling H, Yuan Z, Shen J, et al. Accuracy of matrix-assisted laser desorption ionization-time of flight mass spectrometry for identification of clinical pathogenic fungi: a meta-analysis. J Clin Microbiol 2014;52:2573-82. [PubMed]
- Zhou C, Hu B, Zhang X, et al. The value of matrix-assisted laser desorption/ionization time-of-flight mass spectrometry in identifying clinically relevant bacteria: a comparison with automated microbiology system. J Thorac Dis 2014;6:545-52. [PubMed]
- Rychert J, Burnham CA, Bythrow M, et al. Multicenter evaluation of the Vitek MS matrix-assisted laser desorption ionization-time of flight mass spectrometry system for identification of Gram-positive aerobic bacteria. J Clin Microbiol 2013;51:2225-31. [PubMed]
- Westblade LF, Jennemann R, Branda JA, et al. Multicenter study evaluating the Vitek MS system for identification of medically important yeasts. J Clin Microbiol 2013;51:2267-72. [PubMed]
- Claydon MA, Davey SN, Edwards-Jones V, et al. The rapid identification of intact microorganisms using mass spectrometry. Nat Biotechnol 1996;14:1584-6. [PubMed]
- Holland RD, Wilkes JG, Rafii F, et al. Rapid identification of intact whole bacteria based on spectral patterns using matrix-assisted laser desorption/ionization with time-of-flight mass spectrometry. Rapid Commun Mass Spectrom 1996;10:1227-32. [PubMed]
- Kumar MP, Vairamani M, Raju RP, et al. Rapid discrimination between strains of beta haemolytic streptococci by intact cell mass spectrometry. Indian J Med Res 2004;119:283-8. [PubMed]
- Lau SK, Tang BS, Teng JL, et al. Matrix-assisted laser desorption ionisation time-of-flight mass spectrometry for identification of clinically significant bacteria that are difficult to identify in clinical laboratories. J Clin Pathol 2014;67:361-6. [PubMed]
- Bizzini A, Jaton K, Romo D, et al. Matrix-assisted laser desorption ionization-time of flight mass spectrometry as an alternative to 16S rRNA gene sequencing for identification of difficult-to-identify bacterial strains. J Clin Microbiol 2011;49:693-6. [PubMed]
- Nagy E, Becker S, Kostrzewa M, et al. The value of MALDI-TOF MS for the identification of clinically relevant anaerobic bacteria in routine laboratories. J Med Microbiol 2012;61:1393-400. [PubMed]
- Veloo AC, Knoester M, Degener JE, et al. Comparison of two matrix-assisted laser desorption ionisation-time of flight mass spectrometry methods for the identification of clinically relevant anaerobic bacteria. Clin Microbiol Infect 2011;17:1501-6. [PubMed]
- van Veen SQ, Claas EC, Kuijper EJ. High-throughput identification of bacteria and yeast by matrix-assisted laser desorption ionization-time of flight mass spectrometry in conventional medical microbiology laboratories. J Clin Microbiol 2010;48:900-7. [PubMed]
- Cherkaoui A, Emonet S, Fernandez J, et al. Evaluation of matrix-assisted laser desorption ionization-time of flight mass spectrometry for rapid identification of Beta-hemolytic streptococci. J Clin Microbiol 2011;49:3004-5. [PubMed]


